For your convenience, please use our Oligo Analysis Tool.
The extinction coefficient at 260 nm (e260) is a unique physical property of each oligonucleotide. It is defined as the absorbance at 260 nm of a 1 M aqueous solution measured at 20 °C in an optical cell with 1 cm pathway (Lambert-Beer's law).
Purinic bases show a higher absorption (OD260) than pyrimidinic bases. Interactions between neighbouring bases, as well as modifications that absorb at 260nm, also influence optical absorbance. As a consequence, the extinction coefficient strongly depends on oligonucleotide sequence and composition.
The formula what Eurofins has always used to calculate the amount of substance is:
nmol = OD x 100 / (1.54 x nA + 0.75 x nC + 1.17 x nG + 0.92 x nT)
(
Now, Eurofins has also implemented a new formula to make the calculation of the molar extinction coefficient and thus the determination of amount of substance more accurate. The new formula is based on the nearest-neighbor method and takes into account the sequence specific effects.
nmol = OD x 106 / ε260
Nearest-neighbor and individual-bases extinction coefficients are used in the nearest-neighbors formula when calculating the final, total extinction coefficient of an oligonucleotide:
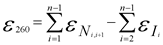
For your oligos you can decide in your personal preferences which formula should be used to calculate the oligo concentration.