You can submit any library pool compatible with Illumina® sequencing, including those prepared from genomic DNA for whole genome sequencing, RNA for transcriptome analysis, or amplicon libraries. Please note, pooling of different libraries into a final ready-to-load pool must be performed by our customers, this service is not offered by Eurofins Genomics.
Illumina-compatible libraries must include four key components in their adapter sequences—located at the ends of each fragment—for successful paired-end sequencing:
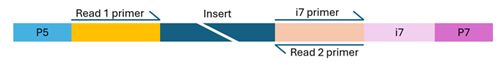
- Flow cell binding sites (P5 and P7): These enable the DNA fragments to attach and cluster on the flow cell.
- Sequencing primer binding sites (Read 1 and Read 2): These sites initiate sequencing from both ends of the DNA fragment.
- Index sequences (i5 and i7): Also known as barcodes, these allow for sample multiplexing. Libraries may be single-indexed (one index) or dual-indexed (two indexes). The usual length of barcodes for NovaSeq is in the range of 8 to 12 bp, while for MiSeq we recommend to use at maximum 10 bp index sequences.
- Index primer binding site(s): These are essential for reading the index sequences during sequencing.
Single-indexed library
Dual-indexed library
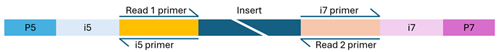
The structure of the adapter sequences varies depending on whether the library uses single or dual indexing. Missing, misconfigured, or heavily modified components can cause sequencing to fail. If you're unsure about your library’s configuration, feel free to contact us for guidance.